Provides age-adjusted incident rates with respect to a reference population, by default seer_std_ages.
Warning! All variables related to your population cohort must be included
in the input data to age_adjust(), otherwise the default assumption will
result in the Florida state population being used for the age-adjustment.
For example if you are calculating incidence rates for subset of counties,
you must include county_name in the input data to age_adjust(), but
county_name should not be a grouping variable. See group_drop() for a
helper function to remove a grouping variable.
age_adjust(data, count = n, population = NULL, population_standard = fcds::seer_std_ages, by_year = "year", age = age_group, keep_age = FALSE)
Arguments
| data | A data frame, containing counts |
|---|---|
| count | The unquoted column name containing raw event or outcome counts. |
| population | Population data specific to the population described by
Note that |
| population_standard | The standard age-specific population used to calculate the age-adjusted rates. By default, uses the 2000 U.S. standard population provided by SEER (see seer_std_ages). |
| by_year | The column or columns by which |
| age | The unquoted column name containing the age or age group. The
default expects that the column |
| keep_age | Age-adjustment by definition summarizes event or outcome
counts (incidence) over all included ages. Set |
| year | The unquoted column name containing the year that will be matched to the population data. |
Value
A data frame with age-adjusted incidence rates in the column rate.
Note that the age column will no longer be included in the output because
the age-adjusted rate summarizes the observed incidence across all ages.
If keep_age is TRUE, the age column is retained, but the final rate is
not calculated, adding the columns population, std_pop, and w for the
specific population, standard population and standardizing population
weight, respectively.
NOTE: The output rate and count are relative to the time span of the
intput data. If the supplied event count data summarizes multiple years --
for example, the FDCS summarized data spans 5 years -- the resulting rate
and count are for the entire period (e.g. 5 years). Convention is to
divide rate by the number of years in the period but leave n as the
raw event count for the period.
Age-Adjusted Rates
Calculating age-adjusted rates requires three primary inputs:
Raw age-specific count of event or outcome, possibly observed or summarized at repeated, consistent time intervals.
Population data with the same demographic resolution as the age-specific counts, as in, for example, the population for the same geographic region, sex, race, year, and age.
The standard reference population that is used to weight incidence among the observed age-specific count.
Each input is required to contain matching age information. The default
data supplied with the package for population (seer_pop_fl) and
population_standard (seer_std_ages) use the column name age_group.
You can specify the name of the column containing age information with the
age argument. If the column name in data is not present in the
population data, age_adjust() will fall back to age_group for those
data sets.
As described in the SEER*Stat Tutorial: Calculating Age-adjusted Rates: The age-adjusted rate for an age group comprised of the ages x through y is calculated using the following formula:
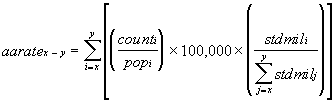
References
https://seer.cancer.gov/seerstat/tutorials/aarates/definition.html
Examples
# This example is drawn from the SEER*Stat Age-adjusted Rate Tutorial: # https://seer.cancer.gov/seerstat/tutorials/aarates/definition.html d_incidence <- dplyr::tribble( ~age_group, ~n, "0 - 4", 116, "5 - 9", 67, "10 - 14", 71, "15 - 19", 87, "20 - 24", 177, "25 - 29", 290, "30 - 34", 657, "35 - 39", 1072, "40 - 44", 1691, "45 - 49", 2428, "50 - 54", 2931, "55 - 59", 2881, "60 - 64", 2817, "65 - 69", 2817, "70 - 74", 2744, "75 - 79", 2634, "80 - 84", 1884, "85+", 1705 ) %>% dplyr::mutate(year = 2013) %>% standardize_age_groups() d_population <- dplyr::tribble( ~age_group, ~population, "0 - 4", 693068, "5 - 9", 736212, "10 - 14", 770999, "15 - 19", 651390, "20 - 24", 639159, "25 - 29", 676354, "30 - 34", 736557, "35 - 39", 724826, "40 - 44", 700200, "45 - 49", 617437, "50 - 54", 516541, "55 - 59", 361170, "60 - 64", 259440, "65 - 69", 206204, "70 - 74", 172087, "75 - 79", 142958, "80 - 84", 99654, "85+", 92692, ) %>% dplyr::mutate(year = 2013) %>% standardize_age_groups() # Because the example data do not include the year of observation, we set # by_year = NULL so that age_adjust() does not attempt to join # d_incidence with d_population by a year column. age_adjust(d_incidence, population = d_population, by_year = NULL)#> # A tibble: 1 x 3 #> n population rate #> <dbl> <dbl> <dbl> #> 1 27069 8796948 400.age_adjust(d_incidence, population = d_population, by_year = NULL, keep_age = TRUE)#> # A tibble: 18 x 6 #> age_group n population std_pop w rate #> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 0 - 4 116 693068 18986520 0.0691 1.16 #> 2 5 - 9 67 736212 19919840 0.0725 0.660 #> 3 10 - 14 71 770999 20056779 0.0730 0.673 #> 4 15 - 19 87 651390 19819518 0.0722 0.964 #> 5 20 - 24 177 639159 18257225 0.0665 1.84 #> 6 25 - 29 290 676354 17722067 0.0645 2.77 #> 7 30 - 34 657 736557 19511370 0.0710 6.34 #> 8 35 - 39 1072 724826 22179956 0.0808 11.9 #> 9 40 - 44 1691 700200 22479229 0.0819 19.8 #> 10 45 - 49 2428 617437 19805793 0.0721 28.4 #> 11 50 - 54 2931 516541 17224359 0.0627 35.6 #> 12 55 - 59 2881 361170 13307234 0.0485 38.7 #> 13 60 - 64 2817 259440 10654272 0.0388 42.1 #> 14 65 - 69 2817 206204 9409940 0.0343 46.8 #> 15 70 - 74 2744 172087 8725574 0.0318 50.7 #> 16 75 - 79 2634 142958 7414559 0.0270 49.7 #> 17 80 - 84 1884 99654 4900234 0.0178 33.7 #> 18 85+ 1705 92692 4259173 0.0155 28.5