Session 5
August 15, 2018
Review
dplyr Homework
Review the homework from Session 4.
You can check out the approach I took in this page. There is more than one way to accomplish each task, so it’s okay if your code looks different!
- What parts of these tasks were difficult?
- Which parts did you find easy?
- What questions did you have as you worked?
- Were you able to find the answers to any of these questions?
Overview
Review
Tidy Data
Two-table dplyr verbs
Tidy Data
Examples
Example 1
There are three variables in this data. What are they?
| Pregnant | Not Pregnant | |
|---|---|---|
| Male | 0 | 5 |
| Female | 1 | 4 |
Tidy…
| sex | pregnant | n |
|---|---|---|
| Male | TRUE | 0 |
| Female | TRUE | 1 |
| Male | FALSE | 5 |
| Female | FALSE | 4 |
Example 2
What are the variables in this data set?
| Patient | 2000 | 2002 | 2004 |
|---|---|---|---|
| John | 10 (Prostate) | 11 (Prostate) | |
| Mary | 9.6 (Breast) | 10.3 (Breast) | 10.5 (Breast) |
Tidy…
| Patient | Year | tumor_size | cancer_type |
|---|---|---|---|
| John | 2000 | NA | NA |
| Mary | 2000 | 9.6 | Breast |
| John | 2002 | 10 | Prostate |
| Mary | 2002 | 10.3 | Breast |
| John | 2004 | 11 | Prostate |
| Mary | 2004 | 10.5 | Breast |
The three rules of tidy data
Columns represent separate variables
Rows represent individual observations
Observational units form tables
Another messy example
What are the variables in this table and how would you transform it into tidy format?
Two-Table dplyr verbs
Much of this material is drawn from Relational Data in the R4DS book.
Example Tables
We have three tables that you can load with
patients
# A tibble: 5 x 3
id name age
<dbl> <chr> <dbl>
1 1 Emmet 63
2 2 Sheilah 61
3 3 Camron 76
4 4 Marisela 58
5 5 Charity 64encounters
# A tibble: 10 x 4
date patient_id blood_test paid
<chr> <int> <lgl> <lgl>
1 2018-08-08 1 TRUE FALSE
2 2018-08-08 3 TRUE TRUE
3 2018-08-09 1 FALSE FALSE
4 2018-08-10 5 TRUE TRUE
5 2018-08-11 3 TRUE FALSE
6 2018-08-13 1 FALSE TRUE
7 2018-08-13 2 TRUE FALSE
8 2018-08-13 5 TRUE FALSE
9 2018-08-13 3 FALSE FALSE
10 2018-08-13 5 FALSE TRUE insurance
# A tibble: 4 x 2
patient_id insurance
<int> <chr>
1 5 Other
2 4 Other
3 1 Aetna
4 2 BCBS Keys
There are two types of keys:
A primary key uniquely identifies an observation in its own table. For example,
patients$idis a primary key because it uniquely identifies each patient in thepatientstable.A foreign key uniquely identifies an observation in another table. For example, the
encounters$patient_idis a foreign key because it appears in thepatientstable where it matches each encounter to a uniquepatient.
What is the primary key of encounters? You can check that a key is primary by counting unique combinations of the variables you expect to be the key.
# A tibble: 5 x 2
# Groups: id [5]
id n
<dbl> <int>
1 1 1
2 2 1
3 3 1
4 4 1
5 5 1# A tibble: 5 x 2
# Groups: date [5]
date n
<chr> <int>
1 2018-08-13 5
2 2018-08-08 2
3 2018-08-09 1
4 2018-08-10 1
5 2018-08-11 1Exercise
What is/are the primary key(s) in the babynames data set?
# A tibble: 10 x 5
year sex name n prop
<dbl> <chr> <chr> <int> <dbl>
1 2011 F Yasemin 14 0.00000724
2 2012 F Madelon 6 0.00000310
3 1956 F Julann 6 0.00000291
4 2006 M Domonique 20 0.00000913
5 1994 M Washington 9 0.00000442
6 1985 F Letha 24 0.0000130
7 2001 F Gala 16 0.00000808
8 1927 M Roosevelt 484 0.000417
9 1995 M Samar 6 0.00000298
10 1999 F Shaelee 16 0.00000822Mutating Joins
A mutating join allows you to combine variables from two tables. It first matches observations by their keys, then copies across variables from one table to the other. http://r4ds.had.co.nz/relation-data.html#mutating-joins
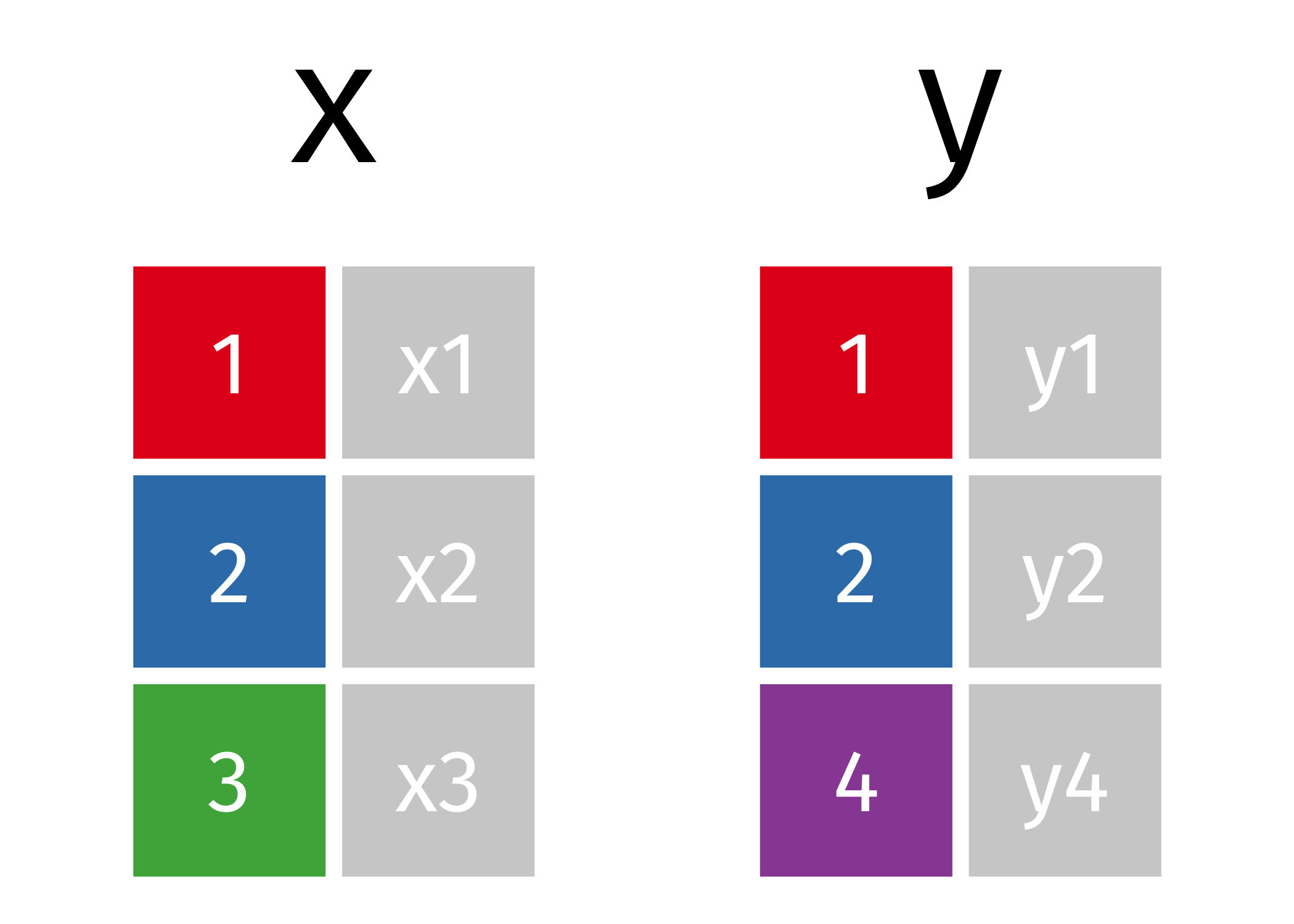
Inner Join
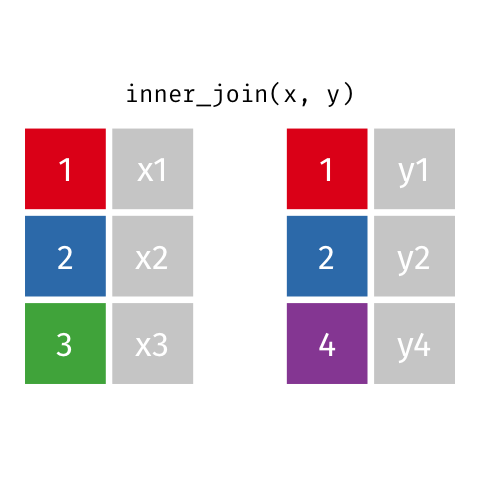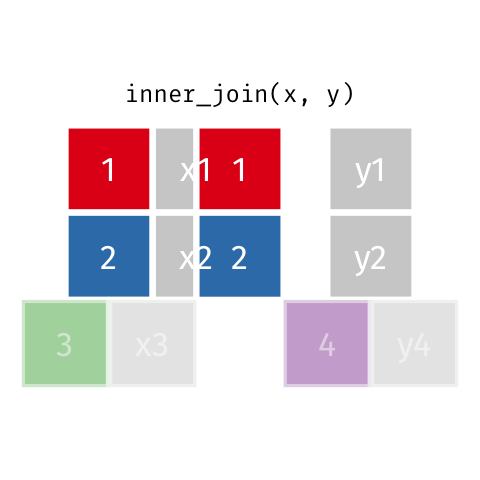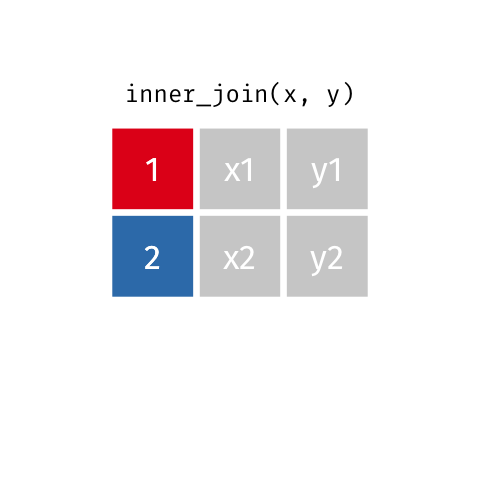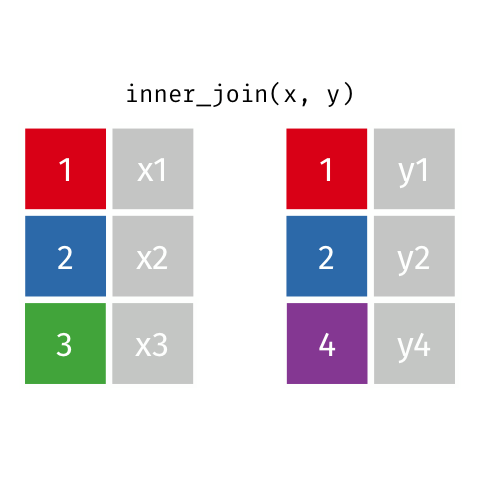
Keeps all observations that appear in
xandy.
# A tibble: 4 x 4
id name age insurance
<dbl> <chr> <dbl> <chr>
1 1 Emmet 63 Aetna
2 2 Sheilah 61 BCBS
3 4 Marisela 58 Other
4 5 Charity 64 Other Left Join
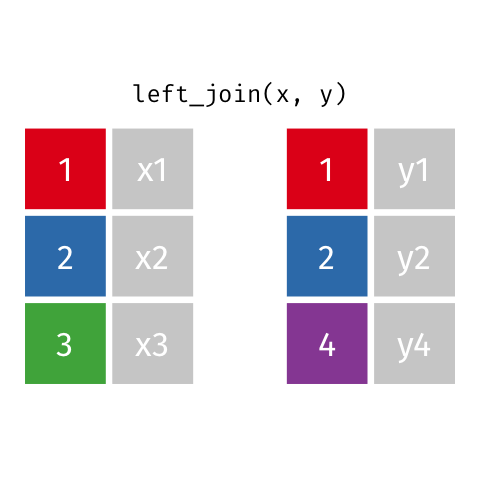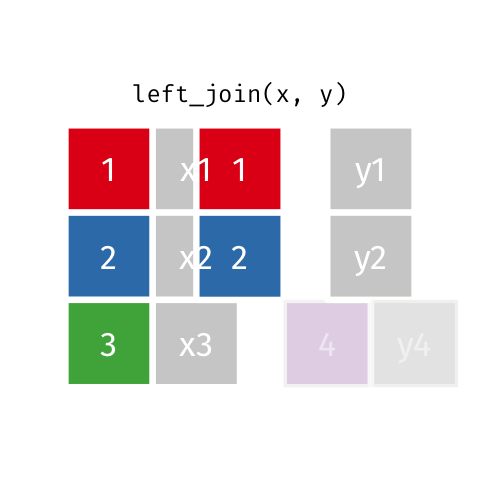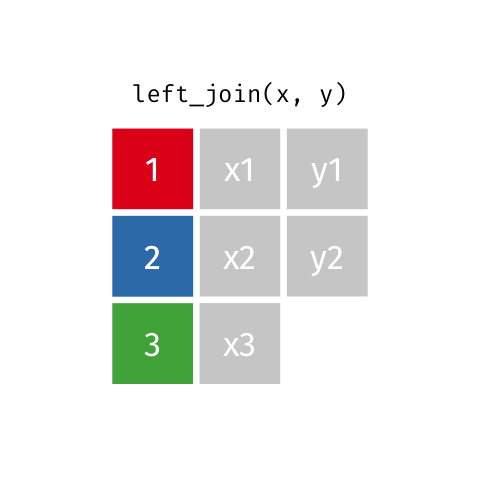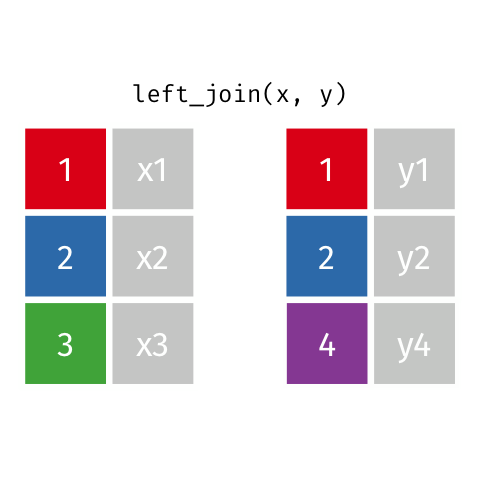
Keeps all observations that appear in
xwith columns inxandy.
# A tibble: 5 x 4
id name age insurance
<dbl> <chr> <dbl> <chr>
1 1 Emmet 63 Aetna
2 2 Sheilah 61 BCBS
3 3 Camron 76 <NA>
4 4 Marisela 58 Other
5 5 Charity 64 Other Left Join 2
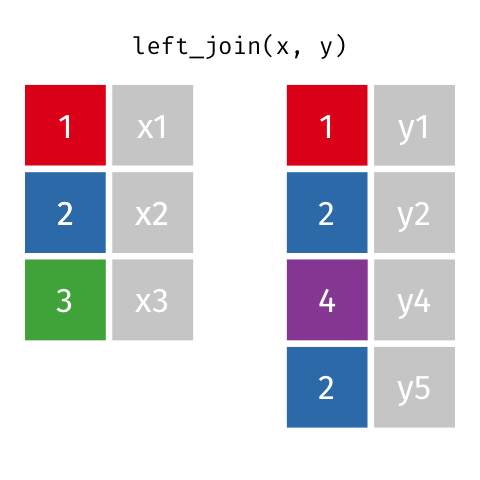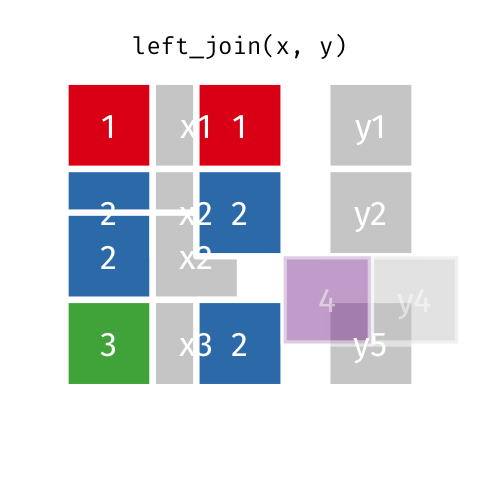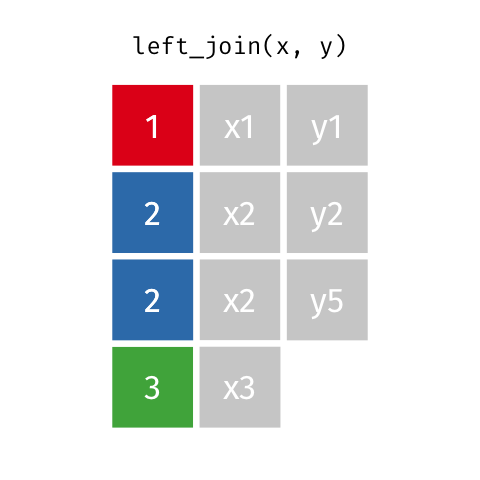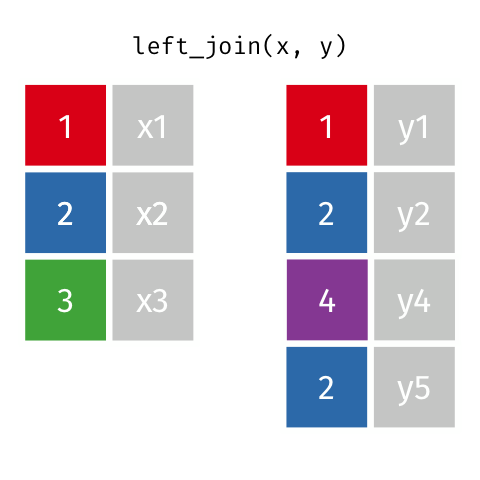
If keys from
xappear multiple times, they appear once for each row iny!
# A tibble: 11 x 6
id name age date blood_test paid
<dbl> <chr> <dbl> <chr> <lgl> <lgl>
1 1 Emmet 63 2018-08-08 TRUE FALSE
2 1 Emmet 63 2018-08-09 FALSE FALSE
3 1 Emmet 63 2018-08-13 FALSE TRUE
4 2 Sheilah 61 2018-08-13 TRUE FALSE
5 3 Camron 76 2018-08-08 TRUE TRUE
6 3 Camron 76 2018-08-11 TRUE FALSE
7 3 Camron 76 2018-08-13 FALSE FALSE
8 4 Marisela 58 <NA> NA NA
9 5 Charity 64 2018-08-10 TRUE TRUE
10 5 Charity 64 2018-08-13 TRUE FALSE
11 5 Charity 64 2018-08-13 FALSE TRUE Right Join
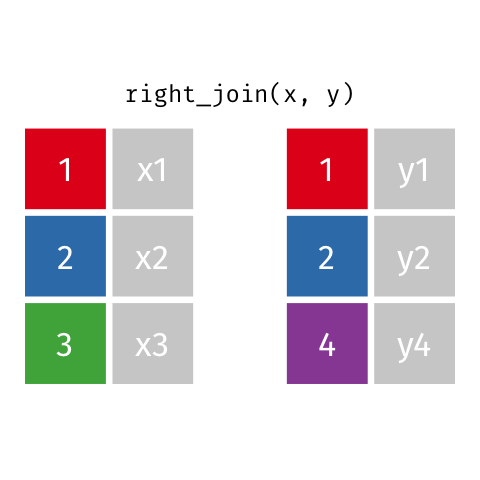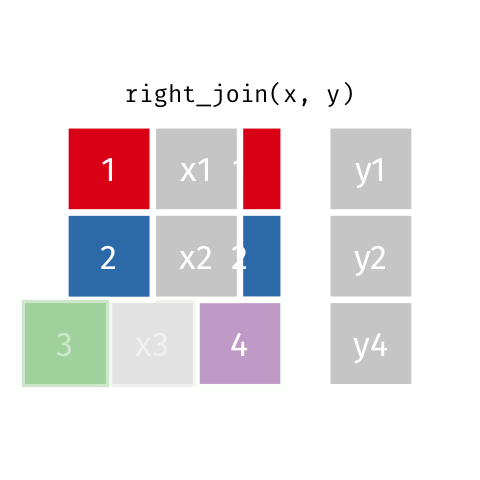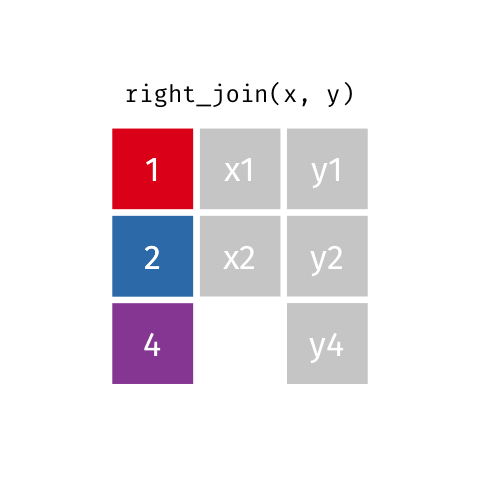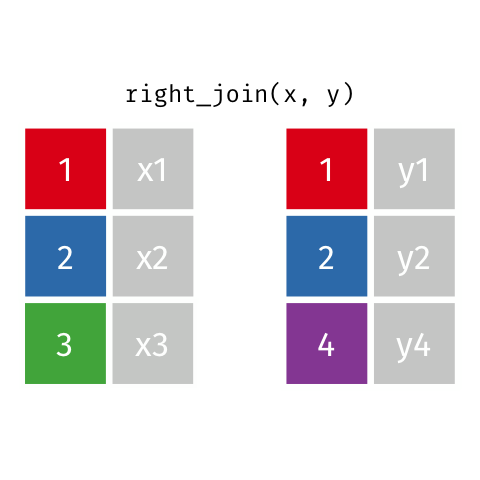
Keeps all observations that appear in
ywith columns inxandy.
# A tibble: 4 x 4
id name age insurance
<dbl> <chr> <dbl> <chr>
1 5 Charity 64 Other
2 4 Marisela 58 Other
3 1 Emmet 63 Aetna
4 2 Sheilah 61 BCBS Full Join
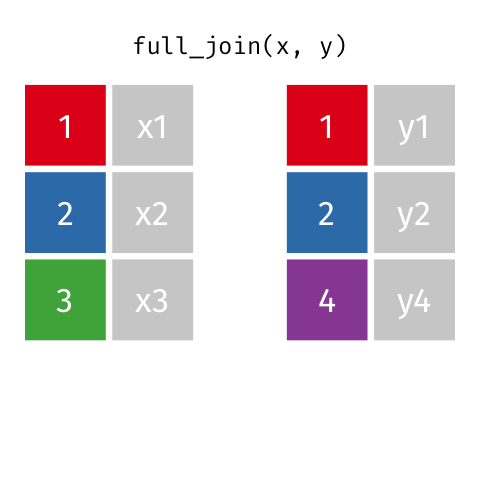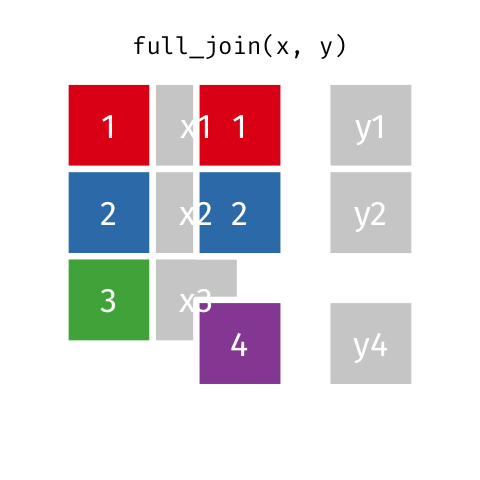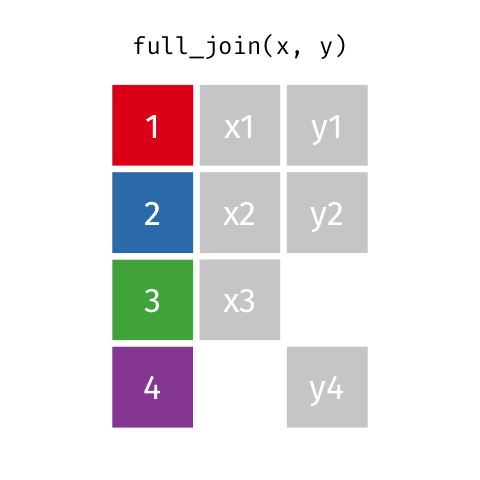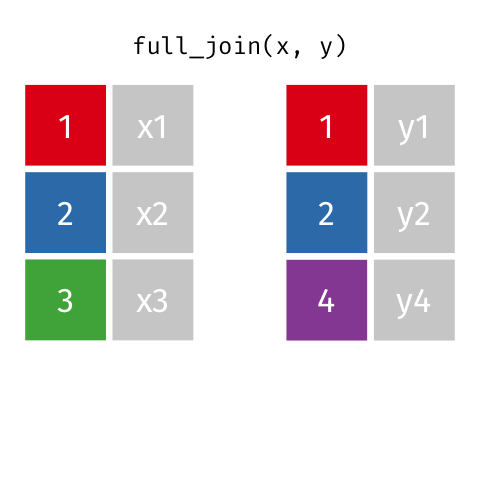
Keeps all observations that appear in
xorywith columns inxandy.
# A tibble: 5 x 4
id name age insurance
<dbl> <chr> <dbl> <chr>
1 1 Emmet 63 Aetna
2 2 Sheilah 61 BCBS
3 3 Camron 76 <NA>
4 4 Marisela 58 Other
5 5 Charity 64 Other Filtering Joins
Filtering joins match observations in the same way as mutating joins, but affect the observations, not the variables.
Semi Join
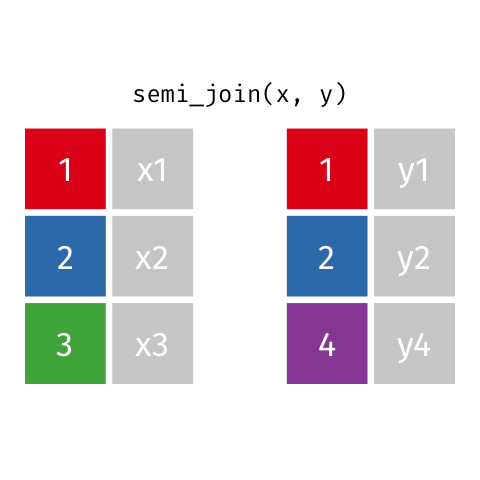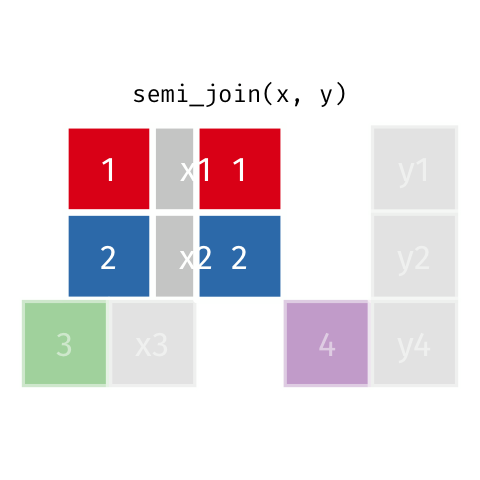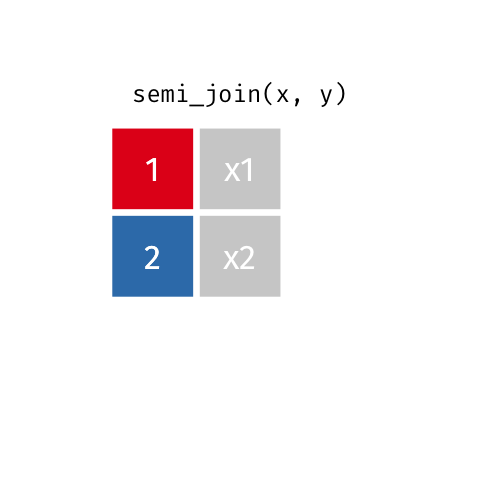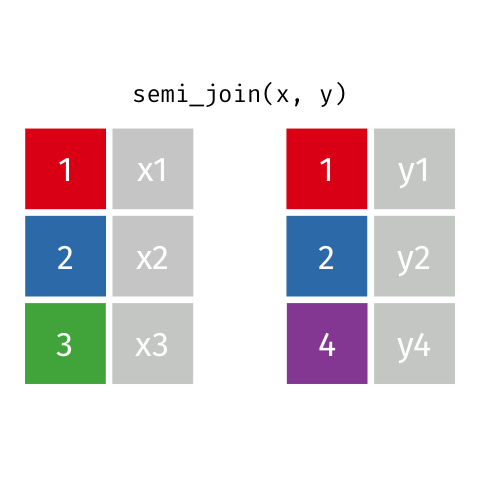
semi_join(x, y)keeps all observations in x that have a match in y.
# A tibble: 4 x 3
id name age
<dbl> <chr> <dbl>
1 1 Emmet 63
2 2 Sheilah 61
3 3 Camron 76
4 5 Charity 64Anti Join
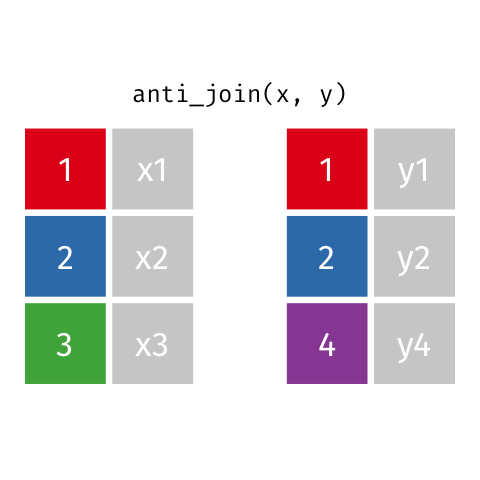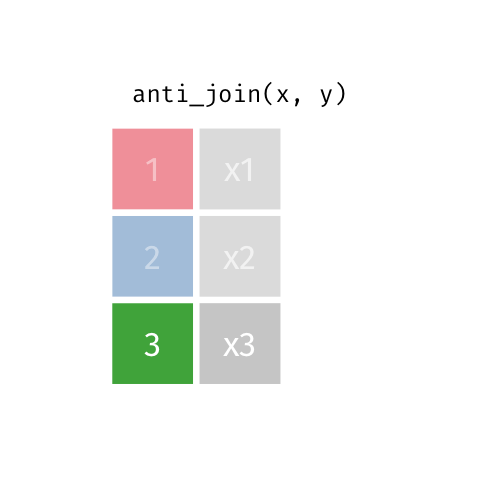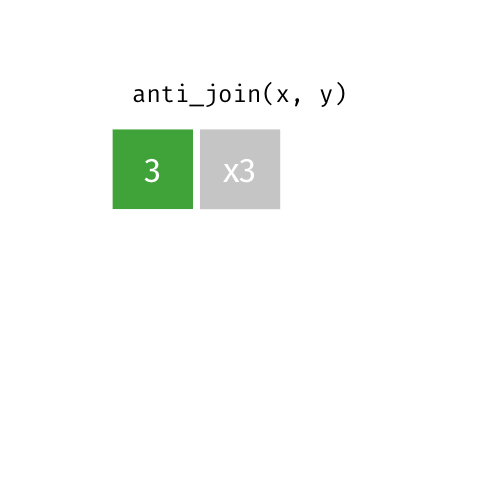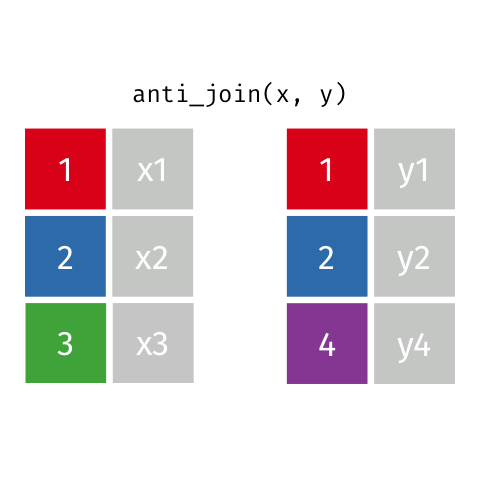
anti_join(x, y)drops all observations in x that have a match in y.
# A tibble: 1 x 3
id name age
<dbl> <chr> <dbl>
1 4 Marisela 58